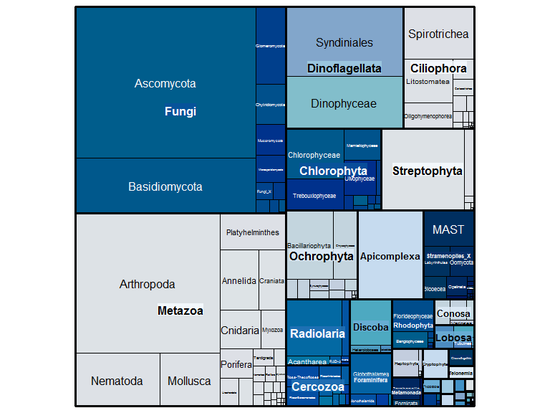
The del Campo Lab studies the ecology and evolution of animals microbiomes and how they are affected by climate change.
About
The lab is led by Dr. Javier del Campo and is part of the Institute of Evolutionary Biology (CSIC-UPF) and the Rosenstiel School of Marine, Atmospheric and Earth Science. The lab is also part of the Diversity of Eukaryotic Microorganisms Consortium (DEMON), a European consortium devoted to the study of the molecular biology, biochemistry, taxonomy, physiology, and ecology of protists from almost all kinds of environments.
Our research of interest includes Microbiology, Ecology and Evolution, which we study using “omics”, imaging and bioinformatics. We are always keen to recruit new lab members if you are interested please contact us.
Principal Investigator
I am a Catalan microbial ecologist and Group Leader at the Institute of Evolutionary Biology (CSIC-UPF) in Barcelona and an Adjunct Professor at the Rosenstiel School of Marine, Atmospheric and Earth Science - University of Miami in Florida. I completed my bachelor’s and my Ph.D. in Environmental Microbiology and Biotechnology at the Institute of Marine Science - CSIC (Barcelona), followed by postdoctoral research at the University of Barcelona, the University of British Columbia (Vancouver), and back to the Institute of Marine Science. In February 2019 I joined as Assistant Professor the Rosenstiel School of Marine and Atmospheric Science at the University of Miami and in February 2021 I joined the Institute for Evolutionary Biology. My research has focused on the study of the ecology and evolution of microbial eukaryotes but recently, I have expanded my scope to the prokaryotes in order to have an integrated view of the microbiome. I am currently investigating microbial community ecology in marine animal-associated environments using cutting-edge sequencing technologies and computational biology. I hope that my research can help to have a better understanding of the role that microorganisms play in the response of animals to climate change.
- Microbial Ecology
- Microbial Evolution
- Microbiome
- Protists
- Climate Change
- eDNA
-
PhD in Environmental Microbiology and Biotechnology, 2011
Universitat de Barcelona and Institut de Ciències del Mar
-
DEA in Environmental Microbiology and Biotechnology, 2005
Universitat de Barcelona
-
BSc in Biology, 2003
Universitat de Barcelona
Projects
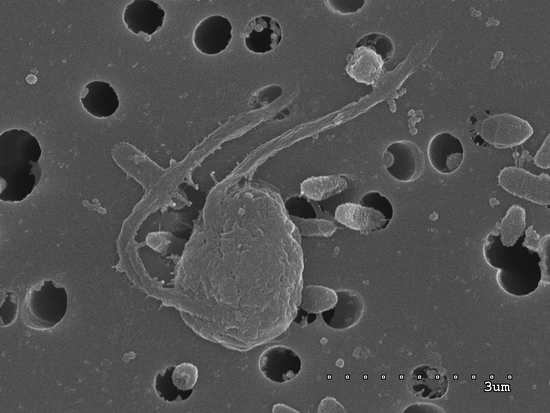
The genome of Mediocremonas mediterraneus
Generation of the reference genome of Mediocremonas mediterraneus as part of the Catalan Biogenome Project

The Montseny Brook Newt Microbiome
Identification and isolation of probiotic bacteria to protect the critically endangered Montseny Brook Newt against chytridiomycosis
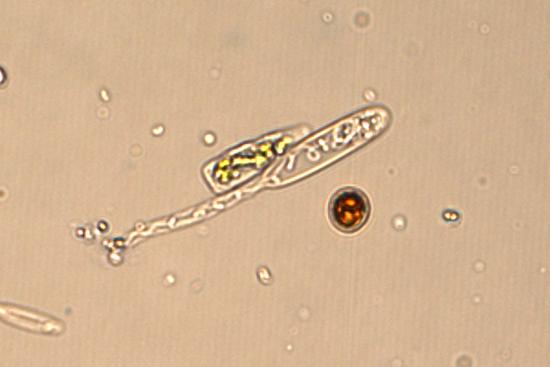
A transcriptomic cell-atlas of reef coral bleaching
Using single-cell transcriptomics to understand the heat stress response of reef coral holobionts.

Biomineralization in the Toadfish microbiome
The microbiome of the Gulf Toadfish (Opsanus beta) and its role in osmoregulation.

Koch at the reef - Understanding coral disease
A systems biology approach to the study of reef corals infectious diseases.
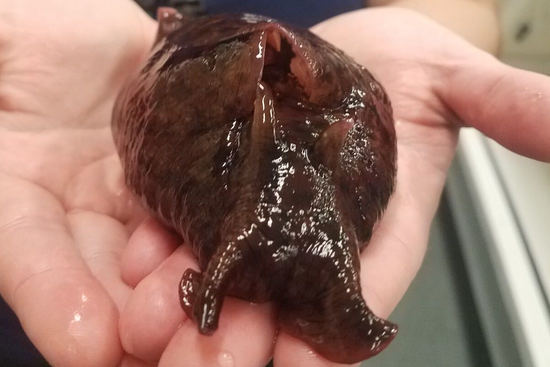
The microbiome of the California sea hare
The microbiome of the neurobiology model organism Aplysia californica.
Recent Publications
Contact
- jdelcampo@ibe.upf-csic.es
- +34 93 542 20 00
- Passeig de la Barceloneta 37-49, Barcelona, Catalonia 08003
- Take the stairs to the second floor and ask, I still don’t know where my office is…
- DM Me
- Skype Me
- Chat on Keybase